Funding for Agri-food Data Canada is provided in part by the Canada First Research Excellence Fund
Research Data Management
In research environments, effective data management depends on clarity, transparency, and interoperability. As datasets grow in complexity and scale, institutions must ensure that research data is FAIR; not only accessible but also well-documented, interoperable, and reusable across diverse systems and contexts in research Data Spaces.
The Semantic Engine (which runs OCA Composer), developed by Agri-Food Data Canada (ADC) at the University of Guelph, addresses this need.
What is the OCA Composer
The OCA Composer is based on the Overlays Capture Architecture (OCA), an open standard for describing data in a structured, machine-readable format. Using OCA allows datasets to become self-describing, meaning that each element, unit, and context is clearly defined and portable.
This approach reduces reliance on separate documentation files or institutional knowledge. Instead, OCA schemas ensure that the meaning of data remains attached to the data itself, improving how datasets are shared, reused, and integrated over time. This makes data easier to interpret for both humans and machines.
The OCA Composer provides a visual interface for creating these schemas. Researchers and data managers can build machine-readable documentation without programming skills, making structured data description more accessible to those involved in data governance and research.
Why Use OCA Composer in your Data Space
Implementing standards can be challenging for many Data Spaces and organizations. The OCA Composer simplifies this process by offering a guided workflow for creating structured data documentation. This can help researchers:
- Standardize data descriptions across projects and teams
- Improve dataset discoverability and interoperability
- Support collaboration through consistent documentation templates (e.g. Data Entry Excel)
- Increase transparency and trust in data definitions
By making metadata a central part of data management, researchers can strengthen their overall data strategy.
Integration and Customization
The OCA Composer can support the creation and running of Data Spaces by organizations, departments, research projects and more. These Data Spaces often have unique digital environments and branding requirements. The OCA Composer supports this through embedding and white labelling features. These allow the tool to be integrated directly into existing platforms, enabling users to create and verify schemas while remaining within the infrastructure of the Data Space. Institutions can also apply their own branding to maintain a consistent visual identity.
This flexibility means the Composer can be incorporated into internal portals, research management systems, or open data platforms including Data Spaces while preserving organizational control and customization.
To integrate the OCA Composer in your systems or Data Space, check out our more technical details. Alternatively, consult with Agri-food Data Canada for help, support or as a partner in your grant application.
Written by Ali Asjad and Carly Huitema
Surprise! Surprise! I’m switching gears a bit for this blog post – off my historical data and data ownership pedestal for a bit 🙂
I want to talk about RDM – Research Data Management – today. For the past decade I’ve been working with colleagues offering workshops on this topic and working with the Research Data Lifecycle – yup also talked about this in the past:
- Research Data Lifecycle (Oct 13, 2023)
- Organizing your Data – RDM (Nov 17, 2023)
- Documenting your work – Variables – RDM (Dec 8, 2023)
- Documenting your work – README – RDM (Jan 5, 2024)
- Documenting your work – Statistical Analysis – RDM (Jan 25, 2024)
Throughout these posts and the FAIR set of blog posts – I always think to myself – we are NOT teaching anyone anything new or really exciting. It is more about bringing these challenges to light and nudging everyone to think about RDM when they start their research projects. At the end of the workshops, I often have students thank me and comment on how funny my examples were and leave. But, once they get more involved with their projects – that’s when I get the OH! I get it now! and they start from scratch and re-organize their files and project folders.
As ADC matures, we are getting calls from projects to help with their RDM – specifically with the “management” aspect of their data. Questions are usually to the effect of: we have terabytes of data – what do we do? This is a basic yet VERY daunting question – let’s be honest! So let’s work through this together and hopefully some of the tips we re-share here will help.
Organizing your Research Project Data
When organizing a project there are many ways to do this. Let’s use my bookshelf in my home office as an example. I can organize my books by author, or I can organize my books by topic, or I can organize my books by frequency of use, or I can organize my books by colour, or…. you get the idea! How I organize the books on my shelf is really a personal choice and based on how “I” use the books. Now, let’s turn to how to organize your project data. Chances are you will have many different views and opinions on how to organize the data. The project team may consider organizing it by date received, or by instrument used to collect the data, or by individual collecting the data, the options are almost endless. In my opinion, there are a couple of ways that I think about it: how the data was collected VS how the data will be used.
In extremely large projects where we have terabytes of data, you should start by asking yourself the very basic question: “How will be use this data? or How do we anticipate using this data?” Organizing the data by animal or plot does NOT make sense if you anticipate working with the data across many dates. So would organizing the data by dates be better? Let’s be honest – there is NO one way or right way for all! But, I HIGHLY recommend you and/or your team spend time working through the best organization for your data.
Let me show you WHY this will save you a LOT of time. Here is an image I took of my files from an old work laptop – eek! 15 years ago! There are a LOT of problems with how I organized my work laptop at that time. Now, if I need to find a presentation I did for a conference in 2009 – I will need to open ALL those powerpoint presentations, review the content, rename and place in a more appropriate directory. Friday_April_11 is in a large unorganized directory just doesn’t work! What if I need to find that historical polling data from the 1950s? I would need to open each Excel file, browse the file to determine whether it is the correct one or not. Psst – it’s the one titled “husbands_fauults_maritalStatus” 😉
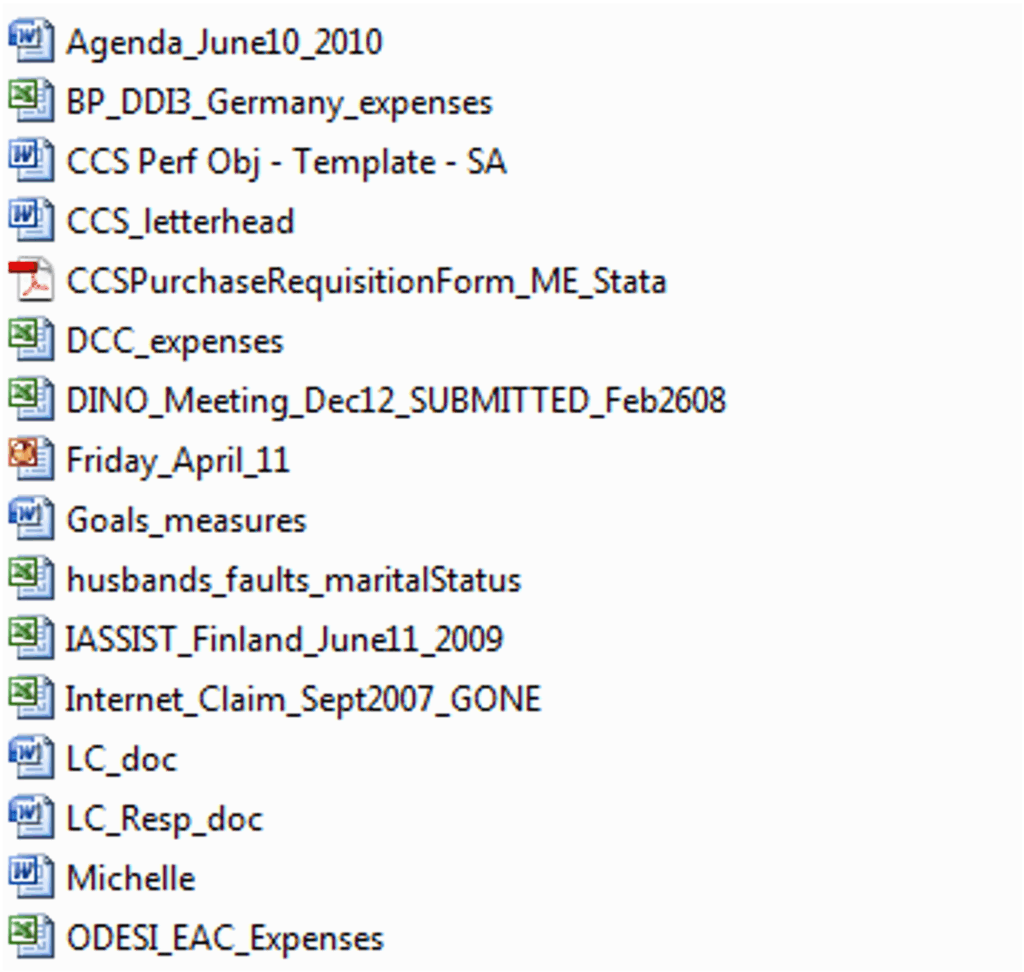
In this directory there are only a few files – so manageable. BUT imagine this was a directory or folder on your computer with Tbs of your data! Names are all over the place, since one instrument may provide filenames as INSTR01.dat, one student may name their files as MEdwards_202505.csv, a third researcher may name their files as PROJECT02_data.xlsx. Without any guidance, everyone places their data files where they think it makes sense – think back to my bookshelf example and you have one big mess – similar to my files back in 2009!
Remember you have Tbs of data! The time it takes to open every file, review the contents, rename, and move to new organizational structure is time saved IF you decide on an organizational structure at the start of your project! YES as project management changes, there may also be a re-org of data – but let’s come up with a structure, document it, and leave for the whole team to use!
Sounds easy right??
If you and your team are starting a project and would like to meet with us to help – please send us an email at adc@uoguelph.ca. We are currently working with a couple of larger projects and would love to help you out too!
![]()
image created by AI
It’s me again! Yup back to that historical data topic too! I didn’t want to leave everyone wondering what I did with my old data – so I thought I’d take you on a tour of my research data adventures and what has happened to all that data.
BSc(Agr) 4th year project data – 1987-1988
Let’s start with my BSc(Agr) data – that image you saw in my last post was indeed part of my 4th year project and a small piece of a provincial (Nova Scotia) mink breeding project: “Estimation of genetic parameters in mink for commercially important traits”. The data was collected over 2 years and YES it is was collected by hand and YES I have it my binder (here in my office). Side note: if you have ever worked with mink – it can take days to smell human after working with them 🙂 Now some of you may be thinking – hang on – breeding project – data in hand – um… how were the farms able to make breeding decisions if I had the data? Did they get a copy of the data?
Remember we are talking 40 years ago – and YES every piece of data that we collected – IF it was relevant for any farm decisions, was photocopied and later entered into a farm management system. So, no management data was lost! However, I took bone diameter measurements, length measures, weights at regular intervals, and many more measures – that frankly were NOT necessary or of interest to management of the animals at that time. Now that data – to me – is valuable!! So – what did I do with it? A few years ago – during some down time – I transcribed it and now I have a series of Excel files with the data. Next question would be – where is the data? Another topic for next blog post 😉
MSc project data – 1989-1990
Moving onto my MSc data – “Estimation of swine carcass composition by video image analysis” (https://bac-lac.on.worldcat.org/oclc/27849855?lang=en). Hang on to your hats for this!!



And you thought handwritten data was bad! Here are all the printouts of my MSc thesis data and hand drawn acetate tracings from a variety of pork cuts! Now what? Remember I keep bringing us back to this concept of historical data. Well, this is to show you that historical data comes in many different formats. Does this have value? Should I do something with this?
Well – you should all know the answer by now :). Yes, a couple of years ago I transcribed the raw data sheets into Excel files. But, those tracings – they’re just hanging around for the moment. I just cannot get myself to throw them out – maybe some day I’ll figure out what to do with them. If you have any suggestions – I would LOVE to hear from you.
Also note, that the manager of the swine unit at that time, kept his own records for management and breeding purposes – this data was only for research purposes.
PhD project data – 1995-1997
So up until now – it took work but I was able to transcribe and re-use my BSc(Agr) and MSc research data. Now the really fun part. Here are another couple of pictures that might take some of you back.
 .
. 
Yup! My whole PhD data was either on these lovely 3.5″ diskettes or on a central server – which is now defunct! Now we might excited and think – hey it’s digital! No need to transcribe! BUT and that’s a VERY LOUD
BUT
These diskettes are 30 years old! Yes I bought a USB disk drive and when I went through these only 3 were readable! and the data on them were in a format that I can no longer read without investing a LOT of time and potentially money!
Now the really sad part – these data were again part of a large rotational breeding program. The manager also kept his own records – but there was SO much valuable data, especially the meat quality side of my trials that were not kept and lost! To this day, I am aware that there were years of data from this larger beef trial that were not kept. It’s really hard to see and know that has happened!
Lessons learned?
Have we really learned anything? For me, personally, these 3 studies, have instilled my desire to save research data – but I have come to realize that not everyone feels the same way. That’s ok! Each of us, needs to consider if there is an impact to losing that OLD or historical data. For my 3 studies, the mink one – the farm managers kept what they needed and the extra measures I was taking would not have impacted the breeding decisions or the industry – so – ok we can let that data die. It’s a great resource for teaching statistics though.
My MSc data – again – I feel that it followed a similar pattern than my BSc(Agr) trials. Although, from a statistical point of view – there are a few great studies that someone could do with this data – so who knows if that will happen or not.
Now my PhD data – that one really stings! Working with the same Research Centre today yes 30 years later! I wish we had a bigger push to save that data. Believe me – we tried – there are a few of us around today that still laugh at the trials and tribulations of creating and resurrecting the Elora Beef Database – but we just haven’t gotten there yet – and I personally am aware of a lot of data that will never be available.
So I ask you – is YOUR research data worth saving? What are your research data adventures? Where will it leave your data?
![]()
image created by AI
I left off my last blog post with a question – well, actually a few questions: WHO owns this data? The supervisor – who is the PI on the research project you’ve been hired onto? OR you as the data collector and analyser? Hmmm…… When you think about these questions – the next question becomes WHO is responsible for the data and what happens to it?
As you already know there really are NO clear answers to these questions. My recommendation is that the supervisor, PI, lab manager, sets out a Standard Operating Procedures (SOP) guide for data collection. Yes, I know this really does NOT address the data ownership question – but it does address my last question: WHO is responsible for the data and what happens to it? And let’s face it – isn’t that just another elephant in the room? Who is responsible for making the research data FAIR?
Oh my, have I just jumped into another rabbit hole?
We have been talking about FAIR data, building tools, and making them accessible to our research community and beyond – BUT? are we missing the bigger vision here? I talk to researchers and most agree that they want to make their data FAIR and share it beyond their lab – BUT…. let’s be honest – that’s a lot of work! Who is going to do it? Here, at ADC, our goal is to work with our research community to help them make agri-food data (and beyond) FAIR – and we’ve been creating tools, creating training materials, and now we are on the precipice of changing the research data culture – well I thought we were – and now I’m left wondering – who is RESPONSIBLE for setting out these procedures in a research project? WHO should be the TRUE force behind changing the data culture and encouraging FAIR research data?
Don’t worry – for anyone reading this – we are VERY set and determined to changing the research data culture by continuing to make the transition to FAIR data – easy and straightforward. It’s just an interesting question and one I would love for you all to consider – WHO is RESPONSIBLE for the data collected in a research project?
Till the next post – let’s consider Copyright and data – oh yes! Let’s tackle that hurdle 🙂
![]()
image created by AI
GitHub is more than just a code repository, it is a a powerful tool for collaborative documentation and standards development. GitHub is an important tool for the development of FAIR data. In the context of writing and maintaining documentation, GitHub provides a comprehensive ecosystem that enhances the quality, accessibility, and efficiency of the process. Here’s why GitHub is invaluable for documentation:
- Version Control: Every change to the documentation is tracked, ensuring that edits can be reviewed, reverted, or merged with ease. This enables a clear history of revisions with clear authorship identified to contributors. While this is possible using a tool such as Google Docs or even Word, version control is a central feature of GitHub and it offers much stronger tooling compared to other methods.
- Collaboration: GitHub makes collaboration easy among team members. Contributors can suggest changes, discuss updates, and resolve questions through pull requests and issues.
- Accessibility: Hosting documentation on GitHub makes it easily accessible to a wide audience. Users can view, clone, or download the latest version of documentation from anywhere.
- Markdown Support: GitHub natively supports Markdown which is a simple and powerful way to create and format documentation. Markdown lets you write clean, readable text with minimal effort.
- Integration and Automation: GitHub integrates with various tools and services. One common usage in documentation is the ability to connect GitHub content with static site generators (e.g., Jekyll, Docusaurus). This then allows documentation to be presented as a webpage with a clean interface for reading, but with the backend tools of GitHub for content management and collaborative creation.
Learn how to start using GitHub
To learn more about how to use GitHub, ADC has contributed content to this online book with introductions to research data management and how to use GitHub for people who write documentation. This project itself is an example of documentation hosted in GitHub and using the static site generator Jekyll to turn back-end markdown pages into an HTML-based webpage.
From the GitHub introduction you can learn about how to navigate GitHub, write in Markdown, edit files and folders, work on different branches of a project, and sync your GitHub work with your local computer. All of these techniques are useful for working collaboratively on documentation and standards using GitHub.
Agri-food Data Canada is a partner in the recently announced Climate Smart Agriculture and Genomics project and is a member of the Data Hub. One of our outputs as part of this team has been the introduction to GitHub documentation.
Written by Carly Huitema
I have been attending industry-focused meetings over the past month and I’m finding the different perspectives regarding agri-food data very interesting and want to bring some of my thoughts to light.
I first want to talk about my interpretation of the academic views. In research and academia we focus on Research Data Management, how and where does data fit into our research. How can and should we make the data FAIR? During a research project, our focus is on what data to collect, how to collect without bias, how to clean it so we can use it for analysis and eventually draw conclusions to our research question. We KNOW what we are collecting since we DECIDE what we’re collecting. In my statistics and experimental design classes, I would caution my students to only collect what they need to answer their research question. We don’t go off collecting data willy-nilly or just because it looks interesting or fun or because it’s there – there is a purpose.
I’ve also been talking about OAC’s 150th anniversary and how we have been conducting research for many years, yet, the data is no where to be found! Ok, yes I am biased here – as I, personally, want to see all that historical data captured, preserved, and accessible if possible – in other words FAIR. But… what happens when you find those treasure troves of older data and there is little to no documentation? Or the principal investigator has passed on? Do you let the data go? Remember, how I love puzzles and I see these treasure troves as a puzzle – but…. When do you decide that it is not worth the effort? How do you decide what stays and what doesn’t? These are questions that data curators around the world are asking – and ones I’ve been struggling with lately. There is NO easy answer!
In academia, we’ve been working with data and these challenges for decades – now let’s turn our attention to the agri-food industry. Here data has also been collected for decades or more as well – but the data could be viewed more as “personal” data. What happened on my fields or with my animals. Sharing would happen by word of mouth, maybe a local report, or through extension work. Today, though, the data being collected on farms is enormous and growing by the minute – dare I say. As a researcher, I get excited at all that information being collected, but… on a more practical basis – the best way to describe this was penned by a colleague who works in industry – as “So What!”.
Goes back to my statement that we should only collect data that is relevant to our research question. The amount and type of data being collected by today’s farming technology is great but – what are we doing with it? Are producers using it? Is anyone using it? I don’t want to bring the BIG question of data ownership here – so let’s stay practical for a moment – and think about WHY is all that data being collected? WHAT is being done with that data? WHO is going to use it? Oh the questions keep coming – but you get the idea!
In the one meeting I attended – the So What question really resonated with me. We can collect every aspect of what is happening with our soils and crops – but if we can’t use the data when we need it and how we need it – what’s the point?
Yes, I’ve been rambling a bit here, trying to navigate this world of data! So many reasons to make data FAIR and just as many reasons to question making it FAIR. Just as a researcher creates a research question to guide their research, I think we all need to consider the W5 of collecting data: WHO is collecting it, WHAT is being collected, WHERE is it being collected and stored, WHEN is it being collected – on the fly or scheduled, WHY this is the big one!!
A lot to ponder here….
![]()
In today’s data-driven research environment, universities face a growing challenge: while researchers excel at pushing the boundaries of knowledge, they often face challenges managing the technology that supports their work.
Many university research teams still operate on isolated, improvised systems for computing and data storage—servers tucked in closets or offices, ad-hoc storage solutions, and no consistent approach to backups or security. These isolated systems may meet immediate needs but often creates inefficiencies, security risks, and lost opportunities for collaboration and innovation.
In this blog series, we’ll explore how research teams at universities in general can benefit by identifying their community commonalities and consolidating their IT infrastructure. A unified system, professionally managed by a dedicated research IT team, brings enhanced security, greater scalability, improved collaboration, and increased efficiency to researchers, allowing them to focus on discovery, not IT overhead. We’ll break down the benefits of this shift and how it can help research institutions thrive in today’s data-intensive landscape. Specifically, we will describe how a Collaborative Research IT Infrastructure might help University of Guelph researchers meet their IT needs while freeing up researchers’ time to focus on their core research objectives rather than the underlying IT infrastructure.
What You Can Expect from This Series
Post 1: The Problem with Doing It All Yourself
We’ll kick off by examining the issues that arise when research teams manage their own IT infrastructure—uncoordinated systems, security vulnerabilities, inefficient storage, and the burden of maintaining it all. We’ll explore the risks and costs of decentralized research IT infrastructure and the toll it takes on research productivity.
Posted on Nov 29 and available here.
Post 2: Why a Shared Infrastructure Makes Sense
Next, we’ll explore the advantages of moving to a shared research compute and storage system. From cost savings to enhanced security and easier scalability, we’ll show how a well-managed, shared resource pool can transform the way researchers handle data, computations, and infrastructure, giving them access to state-of-the-art tools and adding scalability by leveraging idle capacity from other research groups.
Posted on Jan 10 and available here.
Post 3: Debunking the Myths: Research Autonomy in a Shared System
A common concern is that adopting a shared infrastructure means losing control. In this post, we’ll discuss how a collaborative system can actually increase flexibility, offering tailored environments for different research needs, while freeing researchers from the technical burdens of IT management. We’ll also explore how it fosters easier collaboration across departments and institutions.
Posted on May 16 and available here.
Post 4: The Benefits of Shared Storage
Research generates vast amounts of data, and managing it efficiently is key to success. This post will look at how shared storage solutions offer more than just space—providing reliable backups, cost-effective scaling, and multiple storage tiers to meet various research needs, from active datasets to long-term archives.
Posted on Aug 29 and available here.
Post 5: Scaling for the Future: Building a System That Grows with Your Research
As research projects evolve, so do their IT demands. This post will highlight how shared infrastructure offers scalability and adaptability, ensuring that universities can support growing data and computational needs. We’ll also discuss how investing in shared systems today sets universities up to leverage future advancements in research computing.
Post 6: Transitioning to a Shared System: Key Considerations
In our final post, we’ll discuss key considerations for the University of Guelph to explore the move to a shared research compute and storage system. We’ll look at the importance of securing sustainable funding, fostering consensus across departments, and navigating shared governance to ensure all voices are heard. Additionally, we’ll examine how existing organizational structures influence the establishment of dedicated roles for managing this infrastructure. This discussion aims to highlight the factors that can guide a smooth transition toward a collaborative research IT environment.
The Case for Change
By the end of this series, you’ll have a clear understanding of why shared research infrastructure is the future for universities. We’ll show that this approach isn’t just about technology—it’s about improving collaboration, safeguarding data, and ultimately empowering researchers to focus on what really matters: driving innovation. Join us as we explore the journey from siloed systems to shared success.
Written by Lucas Alcantara
Featured picture generated by Pixlr
There is a new feature just released in the Semantic Engine!
Now, after you have written your schema you can use this schema to enter and verify data using your web browser.
Find the link to the new tool in the Quick Link lists, after you have uploaded a schema. Watch our video tutorial on how to easily create your own schema.
Add data
The Data Entry Web tool lets you upload your schema and then you can optionally upload a dataset. If you choose to upload a dataset, remember that Agri-food Data Canada and the Semantic Engine tool never receive your data. Instead, your data is ‘uploaded’ into your browser and all the data processing happens locally.
If you don’t want to upload a dataset, you can skip this step and go right to the end where you can enter and verify your data in the web browser. You add rows of blank data using the ‘Add rows’ button at the bottom and then enter the data. You can hover over the ?’s to see what data is expected, or click on the ‘verification rules’ to see the schema again to help you enter your data.
If you upload your dataset you will be able to use the ‘match attributes’ feature. If your schema and your dataset use the same column headers (aka variables or attributes), then the DEW tool will automatically match those columns with the corresponding schema attributes. Your list of unmatched data column headers are listed in the unassigned variables box to help you identify what is still available to be matched. You can create a match by selecting the correct column name in the associated drop-down. By selecting the column name you can unmatch an assigned match.
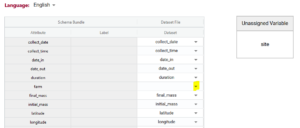
Matching data does two things:
1) Lets you verify the data in a data column (aka variable or attribute) against the rules of the schema. No matching, no verification.
2) When you export data from the DEW tool you have the option of renaming your column names to the schema name. This will automate future matching attempts and can also help you harmonize your dataset to the schema. No matching, no renaming.
Verify data
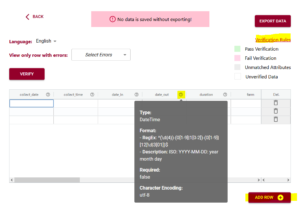
After you have either entered or ‘uploaded’ data, it is time to use one of the important tools of DEW – the verification tool! (read our blog post about why it is verification and not validation).
Verification works by comparing the data you have entered against the rules of the schema. It can only verify against the schema rules so if the rule isn’t documented or described correctly in the schema it won’t verify correctly either. You can always schedule a consultation with ADC to receive one-on-one help with writing your schema.
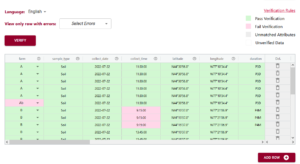
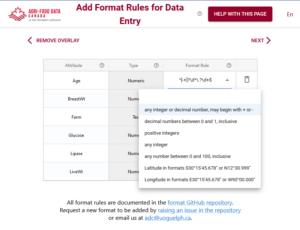
In the above example you can see the first variable/attribute/column is called farm and the DEW tool displays it as a list to select items from. In your schema you would set this feature up by making an attribute a list (aka entry codes). The other errors we can see in this table are the times. When looking up the schema rules (either via the link to verification rules which pops up the schema for reference, or by hovering over the column’s ?) you can see the expected time should be in ISO standard (HH:MM:SS), which means two digits for hour. The correct times would be something like 09:15:00. These format rules and more are available as the format overlay in the Semantic Engine when writing your schema. See the figure below for an example of adding a format rule to a schema using the Semantic Engine.
Export data
A key thing to remember, because ADC and the Semantic Engine don’t ever store your data, if you leave the webpage, you lose the data! After you have done all the hard work of fixing your data you will want to export the data to keep your results.
You have a few choices when you export the data. If you export to .csv you have the option of keeping your original data headers or changing your headers to the matched schema attributes. When you export to Excel you will generate an Excel following our Data Entry Excel template. The first sheet will contain all the schema documentation and then next sheet will contain your data with the matching schema attribute names.
The new Data Entry Web tool of the Semantic Engine can help you enter and verify your data. Reuse your schema and improve your data quality using these tools available at the the Semantic Engine.
Written by Carly Huitema
How should you organize your files and folders when you start on a research project?
Or perhaps you have already started but can’t really find things.
Did you know that there is a recommendation for that? The TIER protocol will help you organize data and associated analysis scripts as well as metadata documentation. The TIER protocol is written explicitly for performing analysis entirely by scripts but there is a lot of good advice that researchers can apply even if they aren’t using scripts yet.
“Documentation that meets the specifications of the TIER Protocol contains all the data, scripts, and supporting information necessary to enable you, your instructor, or an interested third party to reproduce all the computations necessary to generate the results you present in the report you write about your project.” [TIER protocol]
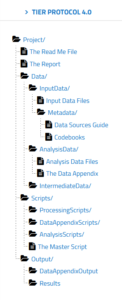
If you go to the TIER protocol website, you can explore the folder structure and read about the contents of each folder. You have folders for raw data, for intermediate data, and data ready for analysis. You also have folders for all the scripts used in your analysis, as well as any associated descriptive metadata.
You can use the Semantic Engine to write the schema metadata, the data that describes the contents of each of your datasets. Your schemas (both the machine-readable format and the human-readable .txt file) would go into metadata folders of the TIER protocol. The TIER protocol calls data schemas “Codebooks”.
Remember how important it is to never change raw data! Store your raw collected data before any changes are made in the Input Data Files folder and never! ever! change the raw data. Make a copy to work from. It is most valuable when you can work with your data using scripts (and stored in the scripts folder of the TIER protocol) rather than making changes to the data directly via (for example) Excel. Benefits include reproducibility and the ease of changing your analysis method. If you write a script you always have a record of how you transformed your data and anyone who can re-run the script if needed. If you make a mistake you don’t have to painstakingly go back through your data and try and remember what you did, you just make the change in the script and re-run it.
The TIER protocol is written explicitly for performing analysis entirely by scripts. If you don’t use scripts to analyze your data or for some of your data preparation steps you should be sure to write out all the steps carefully in an analysis documentation file. If you are doing the analysis for example in Excel you would document each manual step you make to sort, clean, normalize, and subset your data as you develop your analysis. How did you use a pivot table? How did decide which data points where outliers? Why did you choose to exclude values from your analysis? The TIER protocol can be imitated such that all of this information is also stored in the scripts folder of the TIER protocol.
Even if you don’t follow all the directions of the TIER protocol, you can explore the structure to get ideas of how to best manage your own data folders and files. Be sure to also look at advice on how to name your files as well to ensure things are very clear.
Written by Carly Huitema
Findable
Accessible (where possible)
Interoperable
Reusable
I believe most of us are now familiar with this acronym? The FAIR principles published in 2016. I have to admit that part of me really wants to create a song around these 4 words – but I’ll save you all from that scary venture. Seriously though, how many of us are aware of the FAIR principles? Better yet, how many of us are aware of the impact of the FAIR principles? Over my next blog posts we’ll take a look at each of the FAIR letters and I’ll pull them all together with the RDM posts – YES there is a relationship!
So, YES I’m working backwards and there’s a reason for this. I really want to “sell” you on the idea of FAIR. Why do we consider this so important and a key to effective Research Data Management – oh heck it is also a MAJOR key to science today.
R is for Reusable
Reusable data – hang on – you want to REUSE my data? But I’m the only one who understands it! I’m not finished using it yet! This data was created to answer one research question, there’s no way it could be useful to anyone else! Any of these statements sound familiar? Hmmm… I may have pointed some of these out in the RDM posts – but aside from that – truthfully, can you relate to any of these statements? No worries, I already know the answer and I’m not going to ask you to confess to believing or having said or thought any of these. Ah I think I just heard that community sigh of relief 🙂
So let’s look at what can happen when a researcher does not take care of their data or does not put measures into place to make their data FAIR – remember we’re concentrating on the R for reusability today.
Reproducibility Crisis?
Have you heard about the reproducibility crisis in our scientific world? The inability to reproduce published studies. Imagine statements like this: “…in the field of cancer research, only about 20-25% of the published studies could be validated or reproduced…”? (Miyakawa, 2020). How scary is that? Sometimes when we think about reproducibility and reuse of our data – questions that come to mind – at least my mind – why would someone want my data? It’s not that exciting? But boys oh boys when you step back and think about the bigger picture – holy cow!!! We are not just talking about data in our little neck of the woods – this challenge of making your research data available to others – has a MUCH broader and larger impact! 20-25% of published studies!!! and that’s just in the cancer research field. If you start looking into this crisis you will see other numbers too!
So, really what’s the problem here? Someone cannot reproduce a study – maybe it’s age of the equipment, or my favourite – the statistical methodologies were not written in a way the reader could reproduce the results IF they had access to the original data. There are many reasons why a study may not be reproducible – BUT – our focus is the DATA!
The study I referred to above also talks about some of the issues the author encountered in his capacity as a reviewer. The issue that I want to highlight here is access to the RAW data or insufficient documentation about the data – aha!! That’s the link to RDM. Creating adequate documentation about your data will only help you and any future users of your data! Many studies cannot by reproduced because the raw data is NOT accessible and/or it is NOT documented!
Pitfalls to NO Reusable data
There have been a few notable researchers that have lost their career because of their data or rather lack thereof. One notable one is Brian Wansink, formerly of Cornell University. His research was ground-breaking at the time, studying eating habits, looking at how cafeterias could make food more appealing to children, it was truly great stuff! BUT….. when asked for the raw data….. that’s when everything fell apart. To learn more about this situation follow the link I provided above that will take you to a TIME article.
This is a worst case scenario – I know – but maybe I am trying to scare you! Let’s start treating our data as a first class citizen and not an artifact of our research projects. FAIR data is research data that should be Findable, Accessible (where possible), Interoperable, and REUSABLE! Start thinking beyond your study – one never knows when the data you collected during your MSc or PhD may be crucial to a study in the future. Let’s ensure it’s available and documented – remember Research Data Management best practices – for the future.
![]()



