Funding for Agri-food Data Canada is provided in part by the Canada First Research Excellence Fund
Research Data Management
In the rapidly evolving landscape of livestock research, the ability to harness data from diverse sources is paramount. From sensors monitoring animal health to weather data influencing grazing patterns, the insights derived from integrated data can drive informed decisions and innovative solutions. However, integrating data into a centralized livestock research database presents a myriad of challenges that require careful consideration and robust solutions.
Challenges of Data Integration:
- Diverse Data Sources: Livestock research generates data from a multitude of sources, including sensors, health monitoring devices, laboratory tests, and manual observations. Each source may produce data in different formats and structures, complicating the integration process.
- Data Quality and Consistency: Ensuring data quality and consistency across disparate sources is crucial for meaningful analysis and interpretation. Discrepancies in data formats, missing values, and inconsistencies pose significant challenges that must be addressed.
- Real-Time Data Flow: In the dynamic environment of livestock research, timely access to data is essential. Establishing systems for continuous data flow ensures that researchers have access to the latest information for analysis and decision-making.
Solutions for Seamless Data Integration:
- Standardized Data Formats: Implementing standardized data formats, such as JSON or CSV, facilitates easier integration across different sources. By establishing data standards, organizations can streamline the integration process and improve interoperability.
- Data Governance and Quality Assurance: Developing robust data governance policies and quality assurance processes helps maintain data integrity throughout the integration pipeline. Regular audits, validation checks, and data cleaning protocols ensure that only high-quality data is integrated into the research database.
- APIs and Data Pipelines: Leveraging application programming interfaces (APIs) and data pipelines enables automated data retrieval and integration from various sources. APIs provide a standardized way to access and transmit data, while data pipelines automate the flow of data, ensuring seamless integration and synchronization.
- Data Synchronization and Monitoring: Implementing mechanisms for data synchronization and monitoring ensures that data flows continuously and is not missing. Regular checks and alerts can notify database administrators of any disruptions in data flow, allowing for timely resolution.
Conclusion:
In the pursuit of advancing livestock research, data integration plays a pivotal role in unlocking valuable insights and driving innovation. By addressing the challenges associated with integrating data from diverse sources and formats, organizations can create a centralized research database that serves as a foundation for evidence-based decision-making and scientific discovery. Through standardized formats, robust governance practices, and automated data pipelines, seamless data integration becomes achievable, empowering researchers to harness the full potential of data in advancing livestock management and welfare.
Written by Lucas Alcantara
We first talked about writing filenames back in a post about Organizing your data: Research Data Management (RDM)
To further improve your filenaming game, check out a naming convention worksheet from Caltech that helps researchers create a great filenaming system that works with their workflows.
Example: My file naming convention is “SA-MPL-EID_YYYYMMDD_###_status.tif” Examples are “P1-MUS-023_20200229_051_raw.tif” and “P2-DRS-285_20191031_062_composite.tif”.
Why standardize file names?
Standardizing file naming conventions helps researchers better organize their own work and collaborate with others. Here are some key benefits of adopting standardized file naming conventions:
Consistency:
Standardized file naming ensures a consistent structure across files, making it easier for researchers to locate and identify documents. This consistency reduces confusion and streamlines file management.
Improved Searchability:
A standardized naming convention makes searching easier. Users can quickly locate files based on keywords, project names, or other relevant information embedded in the file names, reducing the time spent searching for specific documents.
Ease of Sorting:
Uniform file names make it easier to sort and arrange files in alphabetical or chronological order. This aids in maintaining an organized file structure, especially when dealing with a large volume of documents.
Enhanced Collaboration:
In collaborative environments (such as sharing data with your supervisor), standardized file naming promotes a shared understanding of how files are named and organized.
Version Control:
Including version numbers or dates in file names helps manage version control effectively. This is particularly important in situations where multiple iterations of a document are created, ensuring that users can identify the most recent or relevant version.
By adopting a consistent approach to file naming, researchers can improve their overall file management processes, prevent mistakes, and enhance productivity.
File organization:
Keep reading our blog for an upcoming post on file organization using the TIER protocol.
Written by Carly Huitema
How many of you document your statistical analysis code? SAS and R users, do you add comments in your programs? Or do you fly by the seat of your pants, write, modify code, and know that you’ll remember what you did at a later time? I know we don’t have the time to add all this information in our code, but I cannot stress enough how IMPORTANT it is to do something! I’ll use the same story as I did a few posts ago – and be honest! Will you 100% remember WHY you adjusted that weight variable? HOW you adjusted that weight variable? WHY you dropped that observation or the other? If you’re a paper person like myself – you may have written it down in your lab or notebook. Fabulous! BUT! What happens to your notes when you archive your data and associated statistical programs?
Many if not all of the statistical programs that you are using have the ability to add comments among your code. This is the easiest way to start documenting your statistical analysis. When you archive your data, you will also archive your statistical analysis programs (.sas and .r are text files), so future users of your data will understand how your data was created. Yes, there are other options to capture your documentation, one of these is Markdown. Imagine writing your SAS or R code, adding documentation explaining what you do as you work through the code AND adding documentation to your output – all at the same time!
If you’re curious how Markdown works for both R and SAS, review this tutorial “Saving your code and making your research REPRODUCIBLE”. A more recent workshop demonstrating how to use R Markdown for R processes can be found at the Agri-food Research Data Workshop Series: Workshop 6: Documenting your Data and Processes with R Markdown .
Enjoy! And remember if you have any questions or comments – please let us know at adc@uoguelph.ca
![]()
Maintaining data quality can be a constant challenge. One effective solution is the use of entry codes. Let’s explore what entry codes entail, why they are crucial for clean data, and how they are seamlessly integrated into the Semantic Engine using Overlays Capture Architecture (OCA).
Understanding Entry Codes in Data Entry
Entry codes serve as structured identifiers in data entry, offering a systematic approach to input data. Instead of allowing free-form text, entry codes limit choices to a predefined list, ensuring consistency and accuracy in the dataset.
The Need for Clean Data
Data cleanliness is essential for meaningful analysis and decision-making. Without restrictions on data entry, datasets often suffer from various spellings and abbreviations of the same terms, leading to confusion and misinterpretation.
Practical Examples of Entry Code Implementation
Consider scenarios in scientific research where specific information, such as research locations, gene names, or experimental conditions, needs to be recorded. Entry codes provide a standardized framework, reducing the likelihood of inconsistent data entries.
Overcoming Cleanup Challenges
In the past, when working with datasets lacking entry codes, manual cleanup or tools like Open Refine were essential. Open Refine is a useful data cleaning tool that lets users standardize data after collection has been completed.
Leveraging OCA for Improved Data Management
Overlays Capture Architecture (OCA) takes entry codes a step further by allowing the creation of lists to limit data entry choices. Invalid entries, those not on the predefined list (entry code list), are easily identified, enhancing the overall quality of the dataset.
Language-specific Labels in OCA
OCA introduces a noteworthy feature – language-specific labels for entry codes. In instances like financial data entry, where numerical codes may be challenging to remember, users can associate user-friendly labels (e.g., account names) with numerical entry codes. This ensures ease of data entry without compromising accuracy.
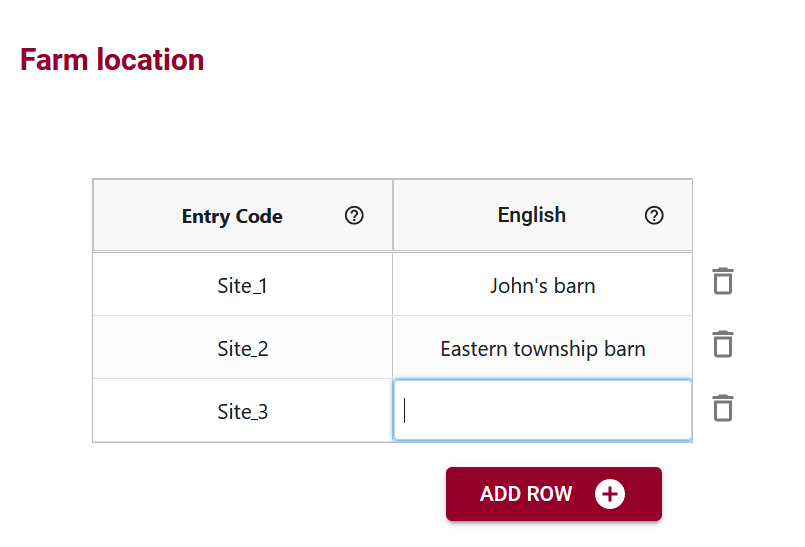
Multilingual Support for Global Usability
OCA’s multilingual support adds a layer of inclusivity, enabling the incorporation of labels in multiple languages. This feature facilitates international collaboration, allowing users worldwide to engage with the dataset in a language they are comfortable with.
Crafting Acceptable Data Entries in OCA
When creating lists in OCA, users define acceptable data entries for specific attributes. Labels accompanying entry codes aid users in understanding and selecting the correct code, contributing to cleaner datasets.
Clarifying the Distinction between Labels and Entry Codes
It’s important to note that, in OCA, the emphasis is on entry codes rather than labels. While labels provide user-friendly descriptions, it is the entry code itself that becomes part of the dataset, ensuring data uniformity.
In conclusion, entry codes play an important role in streamlining data entry and enhancing the quality of datasets. Through the practical implementation of entry codes supported by Overlays Capture Architecture, organizations can ensure that their data remains accurate, consistent, and accessible on a global scale.
Happy New Year everyone!!! Welcome to 2024 – Leap year!!
Oh wow! How time is really flying by! It’s so easy for us to say this and see it happen in our every day lives – BUT – yes it also happens at work and with our research. I remember as a graduate student, in the thick of data collection, thinking I’m never going to finish this project by a given date – there’s too much to do – it’ll never happen! And just like that, whoosh, it’s over and done, and I’ve managed to complete a few research projects since. It’s just amazing how time really does fly by.
As we start a new year with new aspirations, what a great time to implement new habits in our research work! Ah yes, the dreaded documentation piece. Last time we spoke, I talked about variable names and provided you with a list of recommended best practices when creating your variable names for analysis. I also nudged you about keeping those labels, and using the Semantic Engine to create your data schema -check our Carly’s post about Crafting Effective Machine-Actionable Schemas.
So, we have variable names and a data schema, but is that ALL the documentation you should be keeping when you conduct a research project? Of course the answer is NO! Let’s review some other possible documentation pieces and ways to create the documentation.
README file
Let’s tackle the easy piece first and probably the one that will take the longest. A README file is a text file that you should keep in the top folder of your project. Now, let’s first talk about what I mean by a text file. A file created and saved using Notepad on a Windows machine OR TextEdit on a Mac – NOT Word!!! Now I’m sure you’re asking why in the world would I want to use a text editor – a program with NO formatting ability – my document is going to be ugly! Yes it will! BUT – by using a text editor, aka creating a file with a .txt ending will provide you with the comfort that your file will be readable by researchers in the future. Thinking about the Word program as an example, are you 100% positive that the next release will be readable say 5 years from now? Can we read older Word documents today? If you have an older computer with an older version of Word, can you read a document that was created in a newer version of Word? Chances are you’ll have formatting challenges. So…. let’s just avoid that nonsense and use a format that is archivable! .txt
So now that we got that out of the way, what should we include in a README file? Think of the README file as your project or study level documentation. This is where you will describe your folder structure and explain your acronyms. This is also where you will give brief abstract of your study, who the principal investigators are, timeframes, and any information you believe should be passed on to future researchers. Things like challenges with data collection – downpour on day 10 prevented data collection to occur – data collection was conducted by 3 new individuals on day 15, etc… Think about the information that you would find important if YOU were using another study’s data. If you are looking for examples, check out the READMEs in the OAC Historical Research Data and Reproducibility Project dataverse.
The README file is often a skipped yet crucial documentation piece to any project. Some projects use a lab book to capture this information. No matter what media you use the end goal is to capture this information and create a text file for future use.
Conclusion
One more piece of documentation I want to talk about is capturing what happens in your statistical analysis. Let’s leave that for the next post.
![]()
In the dynamic realm of research, where data is the cornerstone of ground-breaking discoveries, ensuring the integrity, reproducibility, and collaboration of your work is paramount. One indispensable tool that researchers often overlook but should embrace wholeheartedly is version control. In this blog post, we’ll delve into the importance of version control for research data, explore its benefits, and provide a few tricks on using GitHub to effectively track changes in your analysis code.
The Importance of Version Control in Research Data
- Preserving Data Integrity
Version control acts as a safety net for your research data, preserving its integrity by keeping a historical record of changes. This ensures that no information is lost or irreversibly altered, allowing researchers to retrace their steps if necessary.
- Enhancing Reproducibility
Reproducibility is a cornerstone of scientific research. Version control enables the precise recreation of experiments and analyses by maintaining a detailed history of code changes. This not only validates your findings but also allows others to replicate and build upon your work.
- Facilitating Collaboration
Collaborative research projects often involve multiple contributors. Version control platforms, such as GitHub, streamline collaboration by providing a centralized hub where team members can contribute, review, and merge changes seamlessly. This collaborative environment fosters a more efficient and organized research process.
- Streamlining Peer Review
Version control systems offer a transparent view of the evolution of your research, making it easier for peers and reviewers to understand and assess your methodology. This transparency enhances the credibility of your work and facilitates a smoother peer-review process.
Tricks for Using GitHub to Track Changes
- Branching Strategies
Leverage GitHub’s branching features to create separate branches for different features or experimental approaches. This allows you to experiment without affecting the main codebase and facilitates the integration of successful changes.
- Commits with Descriptive Messages
Adopt a disciplined approach to commit messages. Each commit should have a clear and concise message describing the changes made. This practice enhances traceability and makes it easier to understand the purpose of each modification.
- Utilize Pull Requests
GitHub’s pull request feature is a powerful tool for code review and collaboration. It enables contributors to propose changes, discuss modifications, and ensure the quality of the code before it is merged into the main branch.
- Continuous Integration
Integrate continuous integration tools with your GitHub repository to automatically test changes and ensure that new code additions do not break existing functionality. This ensures a more robust and error-free codebase.
Learn More with Resources
For those eager to dive deeper into the world of version control and GitHub, there are plenty of accessible resources available. For a comprehensive understanding of version control concepts, “Pro Git” by Scott Chacon and Ben Straub is an excellent e-book available for free online. It covers the basics and advanced features of Git, the underlying technology behind GitHub, making it an invaluable resource for both beginners and experienced users.
YouTube is also a treasure trove of tutorials. The “freeCodeCamp.org” channel offers a series of well-structured videos covering everything from the basics to advanced workflows. The tutorials provide hands-on demonstrations, making it easier for visual learners to grasp the concepts.
If you prefer a more interactive learning experience, GitHub Skills is an online platform offering hands-on courses directly within the GitHub environment. These interactive courses guide you through real-world scenarios, allowing you to practice version control concepts in a practical setting.
Lastly, but not the least, Agri-food Data Canada hosted a hands-on “Introduction to GitHub” workshop this summer as part of the successful Research Data Workshop Series, so make sure to check our slide notes to learn more about GitHub and stay tuned for the next editions of this workshop series.
Remember, mastering version control and GitHub is an ongoing journey, and these resources serve as invaluable companions, whether you’re a novice seeking a solid foundation or an experienced user looking to refine your skills. Happy learning!
In conclusion, version control, especially when harnessed through platforms like GitHub, is not just a technical nicety but a fundamental necessity in the realm of research data. By adopting these practices, researchers can safeguard the integrity of their work, enhance collaboration, and contribute to the broader scientific community with confidence. Embrace version control and watch your research journey unfold with unprecedented clarity and efficiency.
Lucas Alcantara
The next stop on our RDM travels is “Documenting your work”. Those 3 words can scare a lot of people – let’s face it that means spending time writing things down, or creating scripts, or it could be viewed as taking time away from conducting research and analysis. Yes, I know, I know – and anyone who has worked with me in the past, knows that I value documentation VERY highly! Without documentation, your data is valuable to YOU at this moment, but 6 months or 5 years down the road, without documentation it may become useless. On this note, before I start talking about the details of documenting your work, I would like to share the Data Sharing and Management Snafu in 3 Short Acts video. I cannot believe that this video is 10 years old – but it still SO relevant. If you have not seen it, please watch it! It highlights WHY we are talking about RDM – but near the end it deals with our topic today – documenting your data.
Reference: NYU Health Sciences Library. “Data Sharing and Management Snafu in 3 Short Acts” YouTube, 19 Dec 2012, https://www.youtube.com/watch?v=N2zK3sAtr-4.
Variable Names
So let’s talk about variable names for your statistical analyses. Creating variable names is usually done with statistical analyses packages in mind. Let’s be honest we only want to create the variable names once – if we have to rename them – we increase our chances of introducing oddities in our analyses and outputs. Hmm… could I be talking about personal experiences? How many times, in the past, have I fallen into the trap of naming my variables V1, V2, V3, etc… or ME1 or ME_final? It is so easy to fall into these situations especially when we have a deadline looming. So let’s try to build some habits that will help us avoid these situations and help us create data documentation that can eventually be shared and understood by researchers outside of our inner circle. A great place to begin is by reviewing the naming characteristics of the most popular packages used by University of Guelph researchers – based on a survey I conducted in 2017.
Length of Variable Name
SAS: 32 characters long
Stata: 32 characters long
Matlab: 32 characters long
SPSS: 64 bytes long = 64 characters in English or 32 characters in Chinese
R: 10,000 characters long
1st Character of a Variable Name
SAS: MUST be a letter or an underscore
Stata: MUST be a letter or an underscore
Matlab: MUST be a letter
SPSS: MUST be a letter, an underscore or @,#,$
R: No restrictions found
Special Characters in Variable Names
SAS: NOT allowed
Stata: NOT allowed
Matlab: No restrictions found
SPSS: ONLY Period, @ are allowed
R: ONLY Period is allowed
Case in Variable Names
SAS: Mixed case –Presentation only
Stata: Mixed case – Presentation only
Matlab: Case sensitive
SPSS: Mixed case – Presentation only
R: Case sensitive
Recommended Best Practice for Variable Names
Based on the naming characteristics listed above the following is a list of Recommended Best Practices to consider when naming your variables:
- Set Maximum length to 32 characters
- ALWAYS start variable names with a letter
- Numbers can be used anywhere in the variable name AFTER the first character
- ONLY use underscores “_” in a variable name
- Do NOT use blanks or spaces
- Use lowercase
Example Variable Names
Heading in Excel or description of the measure to be taken → variable name to be used in a statistical analysis
Diet A → diet_a
Fibre length in centimetres → fibre_cm
Location of farm → location
Price paid for fleece → price
Weight measured during 2nd week of trial → weight2
Label or description of the variable
Let’s ALWAYS ensure that we are keeping the descriptive part or label for the variable name documented. Check out the Semantic Engine, an easy to use tool to document your dataset!
Conclusion
Variable names are only one piece of the documentation for any study, but it’s usually the first piece we tend to work on as we collect our data or once we start the analysis. Next RDM post I will talk about the other aspects of documentation and present different ways to do it.
![]()
Show of hands – how many people reading this blog know where their current research data can be found on their laptop or computer? No peaking and no searching! Can you tell me where your data is without looking? Let’s be honest now! I suspect a number do know where your data is, but I will also suggest that a number of you do not. When I hold consulting meetings with students and researchers, quite often I get the “just a minute let me find my data.” “oh that’s not the right one” “it should be here, where did it go?” “I made a change last night and I can’t remember what I called it”. Do any other these sound a little familiar? There’s nothing wrong with any of this, I will confess to saying a lot of these myself – and I teach this stuff – you would think I, of all people should know better. But, we’re all human, and when it gets busy or we get so involved with our work, well…. we forget and take shortcuts.
So, what am I going on about? Organizing your data! Let’s take this post to walk through some recommended best practices.
Project acronym
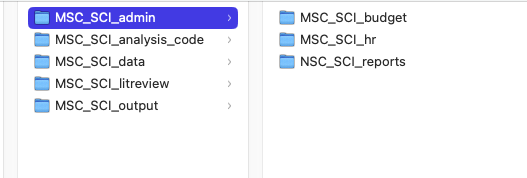
Consider creating an acronym for your project and creating a folder for ALL project information. For example, I have a project working with my MSc data on imaging swine carcasses. I have a folder on my laptop called RESEARCH, then I have a folder for this project called MSC_SCI. Any and all files related to this project can be found in this folder. That’s step one.
Folders
I like to create a folder structure within my project folder. I create a folder for admin, analysis_code, data, litreview, outputs, and anything that seems appropriate to me for the project. Within each of these folders I may create subfolders. For example, under admin, I usually have one for budget, one for hr, and one for reports. Under the data folder I may add subfolders based on my collection procedures.
Take note that all my folders start with my project acronym – an easy way to find the project and all its associated content.
Filenames
This is where the fun begins. A recommended practice is to start all of your filenames with your project acronym. Imagine doing this – whenever you need to find a file – a quick search on your computer for “MSC_SCI” will show all my files! It’s a great step towards organizing your project files. Let’s dig a little further though… What if you came up with a system for your own files where anything that contains data, has the word data in the filename, OR anything dealing with the proposal, has the work proposal in the filename? You see where I’m going right? Yes, your file names will get a little long and this is where you need to manage the length and how much description you keep in the filenames. Recommended filename length is 25 characters, but in the end, it’s up to you how long or how short your filenames are. For us mature researchers, remember the days when all names had to be 8 characters?
Dates
We all love our dates and we tend to include dates in our filenames. Easiest way to determine which was the last file edited, right? How do you add dates though? So many ways and many of them have their own challenges associated with them. The recommendation when you use dates is to use the ISO standard: YYYYMMDD. For example today, the day I am writing this post is November 17, 2023. ISO is 20231117. There is a really cool side effect of creating your dates using the ISO standard – review the next image, can you see what happened?
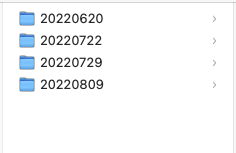
This is an example of a folder where I used the date as the name of the folder that contains data collected on those dates. Notice how they are in order by date collected? Very convenient and easy to see. If I used months spelled out for these dates, I would have August appearing first followed by July and June. If I had other months added, the order, at least to me would be too confusing, as our computers order strings (words) alphabetically. Try the ISO date standard, it takes a bit of getting used to, but trust me you’ll never go back.
Conclusion
Starting a new project with an organized folder structure and a naming convention is a fabulous start to managing your research data. As I say in class and workshops, we are not teaching anything new, we’re encouraging you to implement some of these skills into your research process, to make your life easier throughout your project.
One last note, if you are working in a lab or collaborative situation, consider creating an SOP (Standard Operating Procedure) guide outlining these processes and how you would like to set it up for your lab / group project.
Next stop will be documenting your work.
![]()
Recommendation: Long format of datasets is recommended for schema documentation. Long format is more flexible because it is more general. The schema is reusable for other experiments, either by the researcher or by others. It is also easier to reuse the data and combine it with similar experiments.
Data must help answer specific questions or meet specific goals and that influences the way the data can be represented. For example, analysis often depends on data in a specific format, generally referred to as wide vs long format. Wide datasets are more intuitive and easier to grasp when there are relatively few variables, while long datasets are more flexible and efficient for managing complex, structured data with many variables or repeated measures. Researchers and data analysts often transform data between these formats based on the requirements of their analysis.
Wide Dataset:
Format: In a wide dataset, each variable or attribute has its own column, and each observation or data point is a single row. Repeated measures often have their own data column. This representation is typically seen in Excel.
Structure: It typically has a broader structure with many columns, making it easier to read and understand when there are relatively few variables.
Use Cases: Wide datasets are often used for summary or aggregated data, and they are suitable for simple statistical operations like means and sums.
For example, here is a dataset in wide format. Repeated measures (HT1-6) are described in separate columns (e.g. HT1 is the height of the subject measured at the end of week 1; HT2 is the height of the subject measured at the end of week 2 etc.).
| ID | TREATMENT | HT1 | HT2 | HT3 | HT4 | HT5 | HT6 |
| 01 | A | 12 | 18 | 19 | 26 | 34 | 55 |
| 02 | A | 10 | 15 | 19 | 24 | 30 | 45 |
| 03 | B | 11 | 16 | 20 | 25 | 32 | 50 |
| 04 | B | 9 | 11 | 14 | 22 | 38 | 42 |
Long Dataset:
Format: In a long dataset, there are fewer columns, and the data is organized with multiple rows for each unique combination of variables. Typically, you have columns for “variable,” “value,” and potentially other categorical identifiers.
Structure: It is more compact and vertically oriented, making it easier to work with when you have a large number of variables or need to perform complex data transformations.
Use Cases: Long datasets are well-suited for storing and analyzing data with multiple measurements or observations over time or across different categories. They facilitate advanced statistical analyses like regression and mixed-effects modeling. In Excel you can use pivot tables to view summary statistics of long datasets.
For example, here is some of the same data represented in a long format. The repeated measures don’t have separate columns, compared to the wide format, the height (H) is a column, and the weeks (1-6) are now recorded in a ‘week’ column.
| ID | TREATMENT | WEEK | HEIGHT |
| 01 | A | 1 | 12 |
| 01 | A | 2 | 18 |
| 01 | A | 3 | 19 |
| 01 | A | 4 | 26 |
| 01 | A | 5 | 34 |
| 01 | A | 6 | 55 |
Long format data is a better choice when choosing a format to be documented with a schema as it is easier to document and more clear to understand.
For example, column headers (attributes) in the wide format are repetitive and this results in duplicated documentation. It is also less flexible as each additional week needs an additional column and therefore another attribute described in the schema. This means each time you add a variable you change the structure of the capture base of the schema reducing interoperability.
Documenting a schema in long format is more flexible because it is more general. This makes the schema reusable for other experiments, either by the researcher or by others. It is also easier to reuse the data and combine it with similar experiments.
At the time of analysis, the data can be transformed from long to wide if necessary and many data analysis programs have specialized functions that help researchers with this task.
Written by: Carly Huitema
Anyone who knows me or has sat in one of my classes, will know how much I LOVE the data life cycle. As researchers we have been taught and embraced the research life cycle and I’m sure many of you could recite how that works: Idea → Research proposal → Funding proposal → Data collection → Analysis → Publication → A new idea – and we start again. The data part of this always seemed the part that took the longest – other than maybe the writing – and really just kind of stopped there. As a grad student, many years ago – too many to count anymore – the data was important and I worked with it, massaged it, cleaned it, re-massaged it, analyzed it – until I was happy with the results and my supervisor was happy with the results as well. Then all the work and concentration shifted gears to the chapter writing and publication. The data? Just sat there – with my MSc project the data entry pieces sat in a banker box, until my supervisor cleared out the lab and shipped that box out to me in Alberta or Ontario. So, the data lives, but in a box.
We talk about FAIR data – Findable, Accessible, Interoperable, and Reusable – um…. my MSc data? Is Findable to me – it’s here on the floor under my desk at home. Accessible? maybe -it’s a box of printouts of the raw data that was entered in 1989. Interoperable? Let’s not even think about that! Reusable? um… maybe as a foot stool! So my MSc data as I’m describing it to you right now it NOT FAIR!
Why not? Because we never thought of the data life cycle back then! Collect data, analyze data, publish!
Today, we know better!!! I look back and get sad at the thought of all the data that was collected that well…. no longer is out there – consider my last post about the OAC 150 anniversary?
Today, we strive to observe and follow the data life cycle – we should be telling data’s story – we should be managing our data so that it can be FAIR! Imagine just for a moment, if I had managed my MSc research data – who knows what further research could have been completed. Now, funny story – there was a project here at University of Guelph that was doing what I did with my MSc but with new technologies. The student who worked on the current project reached out to me to talk about my work – all I could do was tell them about my experiences. My data was inaccessible to them – and it turns out so was my thesis – only copy I had was here in my office – and there was/is no accessible PDF version of it. Now – if my data had been managed and archived (I’ll talk more about this in a later post), the student may have been able to incorporate it into her thesis work – now how cool would that have been? Imaging pigs across 30 years? But…. as we know that did not happen.
So I’m going on and on about this – reason is to convince you all – NOT to leave your data to the wayside – you need to manage your research data – you need to create documentation so that YOU can tell your data’s story once you’ve published your work, and so your data can live on, and have the opportunity to play a role in someone else’s project. I never imagined someone else doing similar work than I did 30 years ago – so you just never know!
I’m going to leave this data life cycle diagram above for you to consider. Next time I’ll start digging into the HOWs of Research Data Management (RDM) rather than the WHYs
![]()



