Funding for Agri-food Data Canada is provided in part by the Canada First Research Excellence Fund
Overlays Capture Architecture
In Overlays Capture Architecture (OCA), when using the Semantic Engine you must assign data types to all of your attributes (aka variables). When do you use the array datatype?
You use an array data type when a data record for that attribute would hold multiple values of a specific data type, arranged in a list-like structure. Multiple values is the key. If you perform a measurement, and you record that single value in your data set, that attribute datatype is not an array of values; it is a single value.
However, if you collect multiple measurements, arrange them into a list using a separator to separate each value, and store that list of values in your dataset in a single record for a single attribute (e.g. in a single Excel cell where each value is separated by a comma), then you have an array.
Array data type example
Here are two examples. The table on the left does not have an array data type (it is datatype=numeric) whereas the table on the right contains an array data type (specifically array[numeric]) and uses a comma separator.
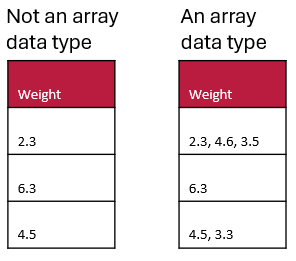
Array data types may be especially useful in questionnaires when you can allow multiple selections for a question (e.g. asking the user to select all the options that apply).
Here are the key characteristics and examples of when you would categorize a data type as an array data type:
- Multiple Elements: An array can store multiple values.
- Same Data Type: All elements in an array must be of the same data type (e.g., all numeric, all strings).
- Indexed Access: Elements in an array can be accessed via their index positions.
In summary, you categorize a data type as an array when it is explicitly defined to contain a collection of elements of the same type, accessible via indices, and useful for storing lists, collections, or sequences of values.
Written by Carly Huitema
Is your data ready to describe using a schema? How can you ensure the fewest hiccups when writing your schema (such as with the Semantic Engine)? What kind of data should you document in your schema and what kinds of data can be left out?
Document data in chunks
When you prepare to describe your data with a schema, try to ensure that you are documenting ‘data chunks’, which can be grouped together based on function. Raw data is a type of data ‘chunk’ that deserves its own schema. If you calculate or manipulate data for presentation in a figure or as a published table you could describe this using a separate schema.
For example, if you take averages of values and put them in a new column and calculate this as a background signal which you then remove from your measurements which you put in another column; this is an summarizing/analyzing process and is probably a different kind of data ‘chunk’. You should document all the data columns before this analysis in your schema and have a separate table (e.g. in a separate Excel sheet) with a separate schema for manipulated data. Examples of data ‘chunks’ include ‘raw data’, ‘analysis data’, ‘summary data’ and ‘figure and table data’. You can also look to the TIER protocol for how to organize chunks of data through your analysis procedures.
Look for Entry Code opportunities
Entry codes can help you streamline your data entry and improve existing data quality.
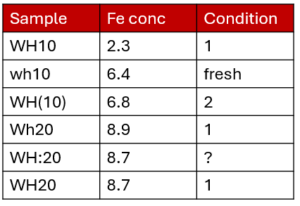
For example, here is a dataset that could benefit from using entry codes. The sample name looks like it would consist of two sample types (WH10 and WH20) but there are multiple ways of writing the sample name. The same thing for condition. You can read our blog post about entry codes which works through the above example. If you have many entry codes you can also import entry codes from other schemas or from a .csv file using the Semantic Engine.
Separate out columns for clarity
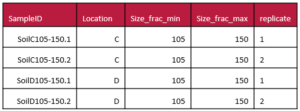
Sometimes you may have compressed multiple pieces of information into a single column. For example, your sample identifier might have several pieces of useful information. While this can be very useful for naming samples, you can keep the sample ID and add extra columns where you pull all of the condensed information into separate attributes, one for each ‘fact’. This can help others understand the information coded in your sample names, and also make this information more easily accessible for analysis. Another good example of data that should be separated are latitude and longitude attributes which benefit from being in separate columns.
Consider adopting error coding
If your data starts to have codes written in the data as you annotate problems with collection or missing samples, consider putting this information in an adjacent data quality column so that it doesn’t interfere with your data analysis. Your columns of data should contain only one type of information (the data), and annotations about the data can be moved to an adjacent quality column. Read our blog post to learn more about adding quality comments to a dataset using the Semantic Engine.
Look for standards you can use
It can be most helpful if you can find ways to harmonize your work with the community by trying to use standards. For example, there is an ISO standard for date/time values which you could use when formatting these kinds of attributes (even if you need to fight Excel to do so!).
Consider schema reuse
Schemas will often be very specific to a specific dataset, but it can be very beneficial to consider writing your schema to be more general. Think about your own research, do you collect the same kinds of data over and over again? Could you write a single schema that you can reuse for each of these datasets? In research schemas written for reuse are very valuable, such as a complex schema like phenopackets, and reusable schemas help with data interoperability improving FAIRness.
In conclusion, you can do many things to prepare your data for documentation. This will help both you and others understand your data and thinking process better, ensuring greater data FAIRness and higher quality research. You can also contribute back to the community if you develop a schema that others can use and you can publish this schema and give it an identifier such as DOI for others to cite and reuse.
Written by Carly Huitema
Entry codes can be very useful to ensure your data is high quality and to catch mistakes that might mess up with your analysis.
For example, you might have taken multiple measurements of two samples (WH10 and WH20) collected during your research.
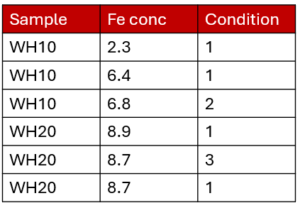
You have a standardized sample name, a measurement (iron concentration) and a condition score for the samples from one to three. You can easily group your analysis into samples because they have consistent names. Incidentally, this is an example of a dataset in a ‘long’ format. Learn more about wide and long formats in our blog post.
The data is clean and ready for analysis, but perhaps in the beginning the data looked more like this, especially if you had multiple people contributing to the data collection:

Sample names and condition scores are inconsistent. You will need to go in and correct the data before you can analyze it (perhaps using a tool such as open refine). Also, if someone else uses your dataset they may not even be aware of the problems, they may not know that the condition score can only have values [1, 2 or 3] and which sample name should be used consistently.
You can help address this problem by documenting this information in a schema using the Semantic Engine with Entry Codes. Look at the figure below to see what entry codes you could use for the data collected.
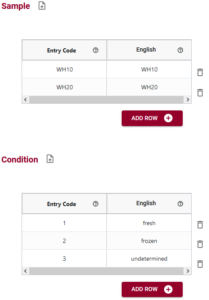
You can see that you have two entry code sets created, one (WH10, WH2) for Sample and one (1, 2, 3) for Condition. It is not always necessary that the labels are different from the entry code itself. Labels become much more important when you are using multiple languages in your schema because it can help with internationalization, or when the label represents some more human understandable concept. In this example we can help understand the Condition codes by providing English labels: 1=fresh sample, 2=frozen sample and 3=unknown condition sample. However, it is very important to note that the entry code (1,2 or 3) and not the label (Fresh, frozen, undetermined) is what appears in the actual dataset.
An example of a complete schema for the above dataset, incorporating entry codes is below:
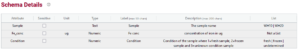
If you have a long list of entry codes you can even import entry codes in a .csv format or from another schema. For example, you may wish to specify a list of gene names and you can go to a separate database or ontology (or use the unique function in Excel) to extract a list of correct data entry codes.
Entry codes can help you keep your data consistent and accurate, helping to ensure your analysis is correct. You can easily add entry codes when writing your schema using the Semantic Engine.
Written by Carly Huitema
Do we verify that the data is correct? Or do we validate it? These two similar terms have different meanings although the two are often conflated.
For data, we can make the following distinction:
Verification: “Did we collect the data right”
Validation: “Did we collect the right data”
For verification we are asking if the data that we collected is consistent with the rules for the form the data should take. For example, if we insist that all the dates must be in the format YYYY-MM-DD, then we can verify the dataset and check that all the dates are indeed in that format. We might also insist on checking that all p-values are between 0 and 1, or that we are using consistent names for all the farms in the dataset.
With the Semantic Engine, you can write the rules needed to verify the dataset right into the schema using overlays such as the format overlay.
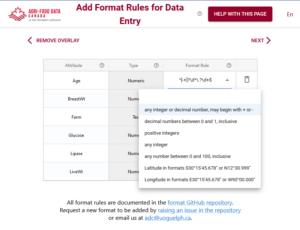
Other rule containing overlays of the schema also contribute verifying a dataset. For example, if a value is required for a specific attribute, then the verification check will make sure there are no blank entries in the dataset for that attribute. If there are entry codes documented in the schema, then data verification will check that only valid entries codes appear in the data for the specific attribute.
Validation is a different beast compared to verification. A researcher collects data to answer a specific research question, such as “Do plants need water to grow”. If you collect data about soil type and plant height and date of death etc. but fail to collect any information about if the plants received water or not, then you are unable to answer the research question. This is of course a trivial example, but it is very easy to get into analysis and realize that you are missing a key variable because you are unable to reach a conclusion. Deciding if your dataset is valid for the research question you are asking is validation and is a key feature of being a researcher.
Typically many people conflate verification and validation, but it becomes very significant especially in a regulatory environment. For example, when the FDA comes and asks for evidence of verification and validation they are asking for very specific things!
The Semantic Engine can help researchers with data verification – did we collect the data right.
Written by Carly Huitema
The Semantic Engine has gotten a recent upgrade for importing entry codes.
If you don’t remember what entry codes are, they help with data standardization and quality by limiting what people can enter in a field for a specific attribute. You can read more about entry codes in our entry code blog post.
Now, the Semantic Engine lets you upload a .csv file that contains your entry codes rather than typing them in individually. You can include the code and multiple languages in your entry code .csv file. Don’t worry if the languages don’t appear in your schema, you will have a chance to pick which ones you want to use.
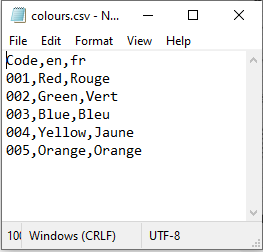
After you have created your .csv file it is time to add them to your schema.
After you have ensured to click ‘list’ for your attribute, it will appear on the screen for adding entry codes. There is an up arrow to select that will let you upload your .csv file containing the entry codes and their labels.
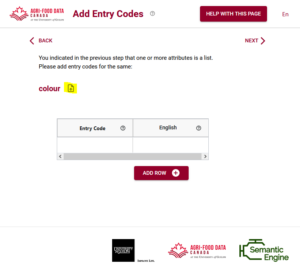
The Semantic Engine will try to auto-match the columns and will give you a screen to check the matching and fill in the correct fields if you need to.
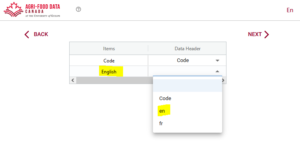
After you have matched columns (and discarded what you don’t need) you now have imported entry codes into your schema. Now you can save the .csv files to reuse when it comes to adding more entry code overlays. It is best practice to include labels for all your languages in your schema, even if they are a repeat of the Code column itself. You can always change it later.
Entry codes are an excellent way to support data quality entry as well as internationalization in your schemas. The Semantic Engine has made it easier to add them using .csv files.
Written by Carly Huitema
You are using a dataset and you come across some missing values, or unusual entries like NA or ND. What do these mean? Why is there data missing?
Quality indicators of individual measurement or observation data can be included directly in your data and will help users of your data understand your data and use it correctly.
Rather than store a quality indicator directly within a data column (such as using a text entry such as NULL or NA in what should be a numerical list of measurements) you should pair your measurement data column with a quality column. This way your measurements will not be ‘contaminated’ with non-numerical indicators and not interfere with analysis.
Documenting your schema with OCA using the Semantic Engine gives you many helpful tools for managing and communicating data quality such as the ability to add entry codes and descriptions to your schema so that others can interpret your data and coding.
To begin with adding quality information you can add an additional column to a measurement column and append _qual to the variable name (or something similar that you recognize and that you document). It is in the ‘var_qual’ column that you can record quality information about the measurements held in the associated ‘var’ column.
Next, to ensure that your ‘var_qual’ column has consistent data entry you can use a system of entry codes. For example, you can set the attribute DataType for the var_qual attribute to be numerical and that it is a list (aka, use entry codes). When it comes to the screen for entering Entry Codes you can enter the following table. You can also adjust the table to be what you need.
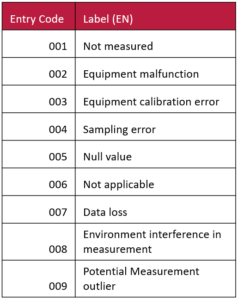
You can also see that you can continue to expand on data quality attributes as you continue your analysis. For example you could create additional quality columns where you might specify the reasons for rejecting values from analysis (e.g. ‘confirmed outlier’ might be one of your entry code labels for this new quality column). Schemas for different stages of collection and analsysi would fit into your data organization structure, such as within the codebook folders of the TIER Protocol.
An example of a dataset that scores the quality of data would be as follows:
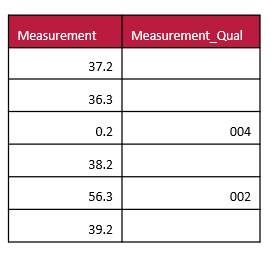
Numbers that are unusual because of equipment errors or other reasons can be flagged and dealt with appropriately during the data analysis cycles.
Written by Carly Huitema
When you document your data schema using the Semantic Engine, you are writing a schema in the schema language of Overlays Capture Architecture (OCA). Let’s do a deeper dive into one of the features of OCA.
Attributes in OCA
When you start using the Semantic Engine, you can either drag and drop your dataset, or you can begin to manually add attributes. Attributes are the names of your columns in your dataset, which should match your variable names in your experiments. In fact, when you drag a dataset into the Semantic Engine, the engine reads the first line of your data, assumes they are your column headers and uses them to create the list of attributes in your schema.
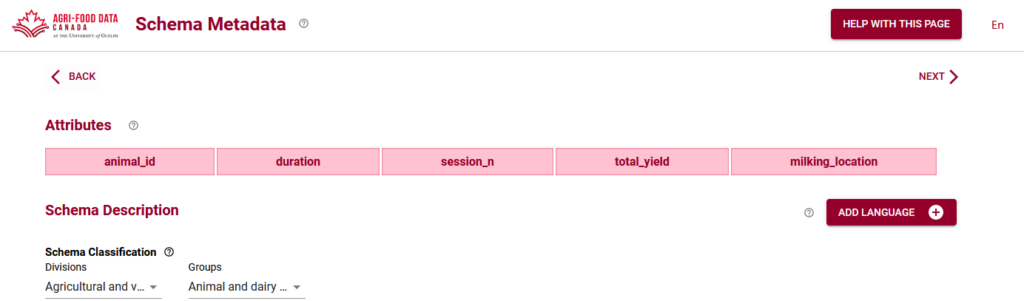
If you are entering your attributes manually, they should be matching the column headers of your dataset.
Labels in OCA
A few screens into the Semantic Engine and you will be asked to add language specific labels. This might be a bit confusing if you’ve already entered in your attributes and you only have one language! The attribute labels and the English language labels might even look exactly the same – and this is OK!
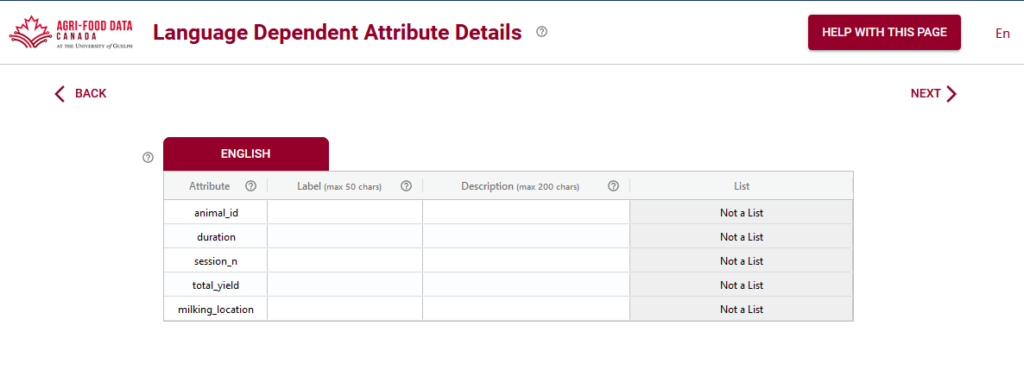
The attributes and their corresponding labels may be the same (or very close), but sometimes they can be very different, especially if the column names are very cryptic. This is your chance to give your data more human readable labels while still preserving the underlying data structure.
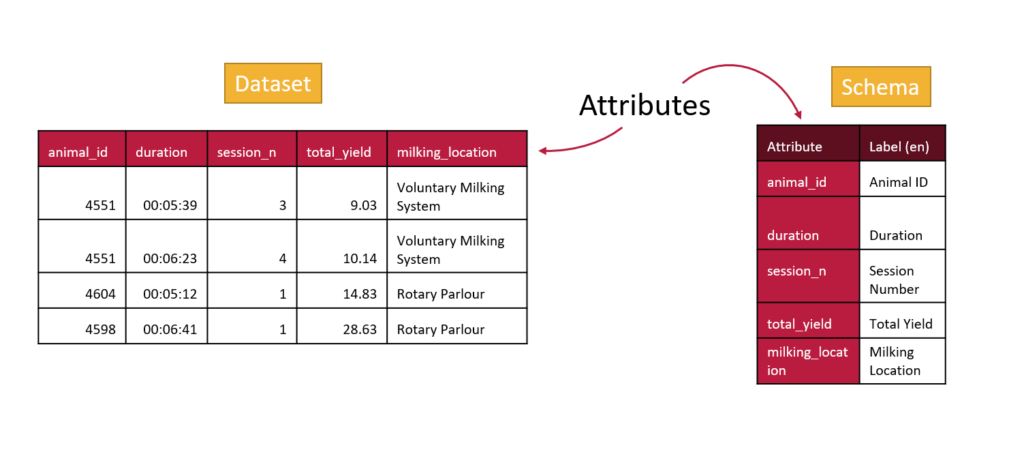
Internationalization with OCA
With labels as well as attributes OCA is able to support internationalization. This means that many people can use the same schemas but they can have helpful information provided to them in their own language.
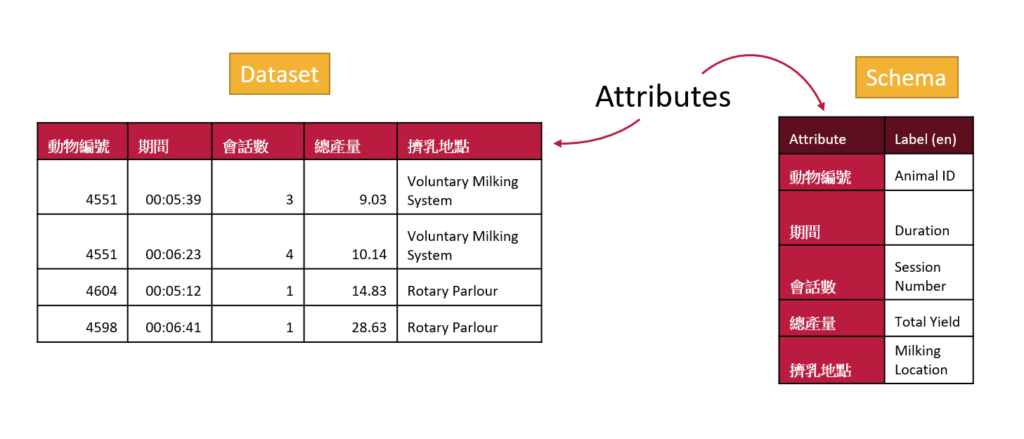
Your labels may not be very different than your attribute names if they are both in English, but the ability to give labels to attributes will help make your schema more accessible in other languages. All you will need to do is edit your schema in the Semantic Engine and add additional languages.
Written by Carly Huitema
Maintaining data quality can be a constant challenge. One effective solution is the use of entry codes. Let’s explore what entry codes entail, why they are crucial for clean data, and how they are seamlessly integrated into the Semantic Engine using Overlays Capture Architecture (OCA).
Understanding Entry Codes in Data Entry
Entry codes serve as structured identifiers in data entry, offering a systematic approach to input data. Instead of allowing free-form text, entry codes limit choices to a predefined list, ensuring consistency and accuracy in the dataset.
The Need for Clean Data
Data cleanliness is essential for meaningful analysis and decision-making. Without restrictions on data entry, datasets often suffer from various spellings and abbreviations of the same terms, leading to confusion and misinterpretation.
Practical Examples of Entry Code Implementation
Consider scenarios in scientific research where specific information, such as research locations, gene names, or experimental conditions, needs to be recorded. Entry codes provide a standardized framework, reducing the likelihood of inconsistent data entries.
Overcoming Cleanup Challenges
In the past, when working with datasets lacking entry codes, manual cleanup or tools like Open Refine were essential. Open Refine is a useful data cleaning tool that lets users standardize data after collection has been completed.
Leveraging OCA for Improved Data Management
Overlays Capture Architecture (OCA) takes entry codes a step further by allowing the creation of lists to limit data entry choices. Invalid entries, those not on the predefined list (entry code list), are easily identified, enhancing the overall quality of the dataset.
Language-specific Labels in OCA
OCA introduces a noteworthy feature – language-specific labels for entry codes. In instances like financial data entry, where numerical codes may be challenging to remember, users can associate user-friendly labels (e.g., account names) with numerical entry codes. This ensures ease of data entry without compromising accuracy.
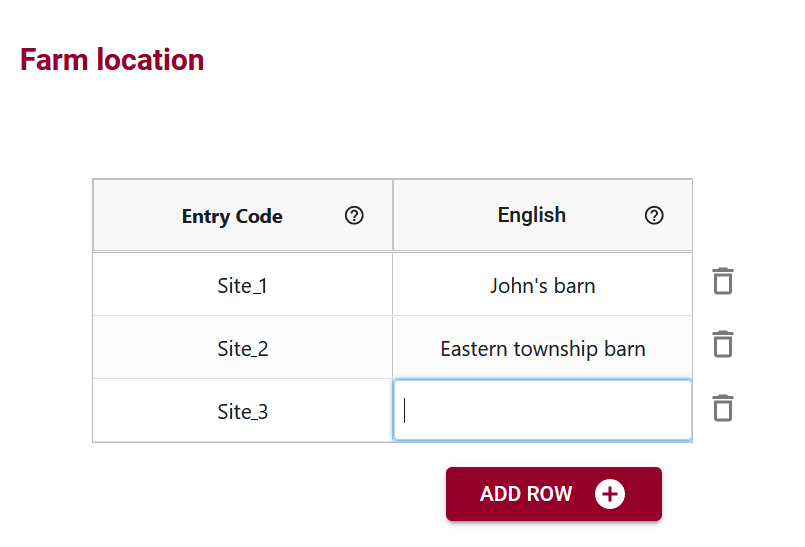
Multilingual Support for Global Usability
OCA’s multilingual support adds a layer of inclusivity, enabling the incorporation of labels in multiple languages. This feature facilitates international collaboration, allowing users worldwide to engage with the dataset in a language they are comfortable with.
Crafting Acceptable Data Entries in OCA
When creating lists in OCA, users define acceptable data entries for specific attributes. Labels accompanying entry codes aid users in understanding and selecting the correct code, contributing to cleaner datasets.
Clarifying the Distinction between Labels and Entry Codes
It’s important to note that, in OCA, the emphasis is on entry codes rather than labels. While labels provide user-friendly descriptions, it is the entry code itself that becomes part of the dataset, ensuring data uniformity.
In conclusion, entry codes play an important role in streamlining data entry and enhancing the quality of datasets. Through the practical implementation of entry codes supported by Overlays Capture Architecture, organizations can ensure that their data remains accurate, consistent, and accessible on a global scale.
When you use the Semantic Engine to create a schema, one of the first things you are asked to do is to classify your schema.
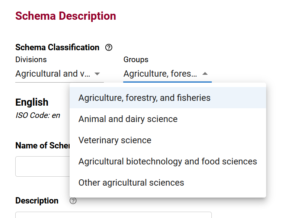
It might seem simple, but as you move further from your domain, what seems like an obvious classification to you may not be so obvious to people outside of your specialty. For example, if someone is talking about a bar there are multiple meanings depending on the context. It could be the location for socialization and drinking, or it could be the exam for lawyers.
With the addition of machine learning and machine assisted searching it is even more important to add important contextual queues to our information to help machines produce more reasonable responses.
A recent publication from the Canadian Federated Research Data Repository (FRDR) demonstrated the challenge they had with automated metadata (e.g. classification) reconciliation. A working group investigated how to build an automated or semi-automated workflow to reconcile metadata keywords from harvested datasets. The majority of their term reconciliation work could not be automated. Ultimately FRDR chose to abandon the assignment of standardized terms to metadata records. The downstream impact means relevant datasets may not appear in relevant searches and research will miss out on opportunities to find and potentially reuse data.
The Semantic Engine supports the findability and categorization of schemas through the addition of schema classifications using the controlled vocabulary of Statistics Canada, specifically the Canadian Research and Development Classification (CRDC) 2020 Version 1.0 – Field of Research (FOR). When you enter your schema classification you are using one of the terms from this controlled list.
Ultimately, by classifying your schema you help ensure that both machines and people can better understand and find your schema and be more confident that they are using it for its intended purpose.
Written by Carly Huitema
Recommendation: Long format of datasets is recommended for schema documentation. Long format is more flexible because it is more general. The schema is reusable for other experiments, either by the researcher or by others. It is also easier to reuse the data and combine it with similar experiments.
Data must help answer specific questions or meet specific goals and that influences the way the data can be represented. For example, analysis often depends on data in a specific format, generally referred to as wide vs long format. Wide datasets are more intuitive and easier to grasp when there are relatively few variables, while long datasets are more flexible and efficient for managing complex, structured data with many variables or repeated measures. Researchers and data analysts often transform data between these formats based on the requirements of their analysis.
Wide Dataset:
Format: In a wide dataset, each variable or attribute has its own column, and each observation or data point is a single row. Repeated measures often have their own data column. This representation is typically seen in Excel.
Structure: It typically has a broader structure with many columns, making it easier to read and understand when there are relatively few variables.
Use Cases: Wide datasets are often used for summary or aggregated data, and they are suitable for simple statistical operations like means and sums.
For example, here is a dataset in wide format. Repeated measures (HT1-6) are described in separate columns (e.g. HT1 is the height of the subject measured at the end of week 1; HT2 is the height of the subject measured at the end of week 2 etc.).
| ID | TREATMENT | HT1 | HT2 | HT3 | HT4 | HT5 | HT6 |
| 01 | A | 12 | 18 | 19 | 26 | 34 | 55 |
| 02 | A | 10 | 15 | 19 | 24 | 30 | 45 |
| 03 | B | 11 | 16 | 20 | 25 | 32 | 50 |
| 04 | B | 9 | 11 | 14 | 22 | 38 | 42 |
Long Dataset:
Format: In a long dataset, there are fewer columns, and the data is organized with multiple rows for each unique combination of variables. Typically, you have columns for “variable,” “value,” and potentially other categorical identifiers.
Structure: It is more compact and vertically oriented, making it easier to work with when you have a large number of variables or need to perform complex data transformations.
Use Cases: Long datasets are well-suited for storing and analyzing data with multiple measurements or observations over time or across different categories. They facilitate advanced statistical analyses like regression and mixed-effects modeling. In Excel you can use pivot tables to view summary statistics of long datasets.
For example, here is some of the same data represented in a long format. The repeated measures don’t have separate columns, compared to the wide format, the height (H) is a column, and the weeks (1-6) are now recorded in a ‘week’ column.
| ID | TREATMENT | WEEK | HEIGHT |
| 01 | A | 1 | 12 |
| 01 | A | 2 | 18 |
| 01 | A | 3 | 19 |
| 01 | A | 4 | 26 |
| 01 | A | 5 | 34 |
| 01 | A | 6 | 55 |
Long format data is a better choice when choosing a format to be documented with a schema as it is easier to document and more clear to understand.
For example, column headers (attributes) in the wide format are repetitive and this results in duplicated documentation. It is also less flexible as each additional week needs an additional column and therefore another attribute described in the schema. This means each time you add a variable you change the structure of the capture base of the schema reducing interoperability.
Documenting a schema in long format is more flexible because it is more general. This makes the schema reusable for other experiments, either by the researcher or by others. It is also easier to reuse the data and combine it with similar experiments.
At the time of analysis, the data can be transformed from long to wide if necessary and many data analysis programs have specialized functions that help researchers with this task.
Written by: Carly Huitema



